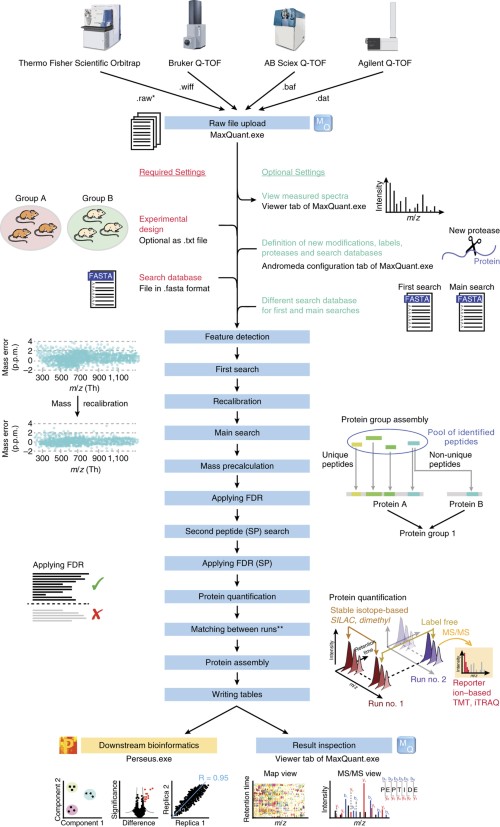
43 maxquant label free quantification
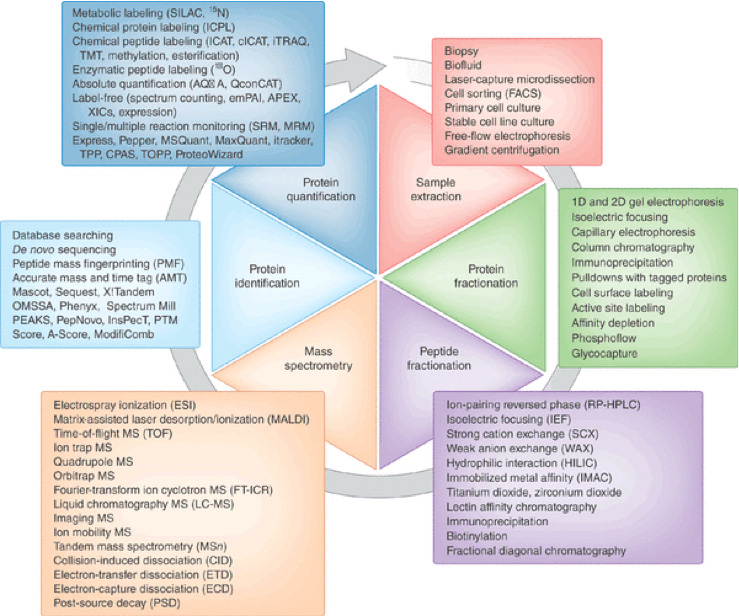
Label Free Quantitation using Maxquant LFQ (MaxLFQ) MaxLFQ is a generic label-free quantification technology that is readily applicable to many biological questions; it is compatible with standard statistical analysis workflows, and it has been validated in many and diverse biological projects. Our algorithms can handle very large experiments of 500+ samples in a manageable computing time. Label-Free Quantification - an overview | ScienceDirect Topics label-free quantification is a method in ms that determines the relative amount of proteins in two or more biological samples, but unlike other quantitative methods, is does not use a stable isotope that chemically binds and labels the protein.70 typically, peptide signals are detected at the ms1 level, and their isotopic pattern allows …
PDF Label-free Quantification with PEAKS - Bioinformatics Solutions Inc. Label-free Quantification with PEAKS Wen Zhang, Ziaur Rahman, Yifang Chen, Lei Xin, and Baozhen Shan Bioinformatics Solutions Inc., Waterloo, Canada Aims Label-free quantification (LFQ) data analysis Summary: ... MaxQuant between the neighboring groups were shown in Figure 2. The UPS1 protein ratios

Maxquant label free quantification
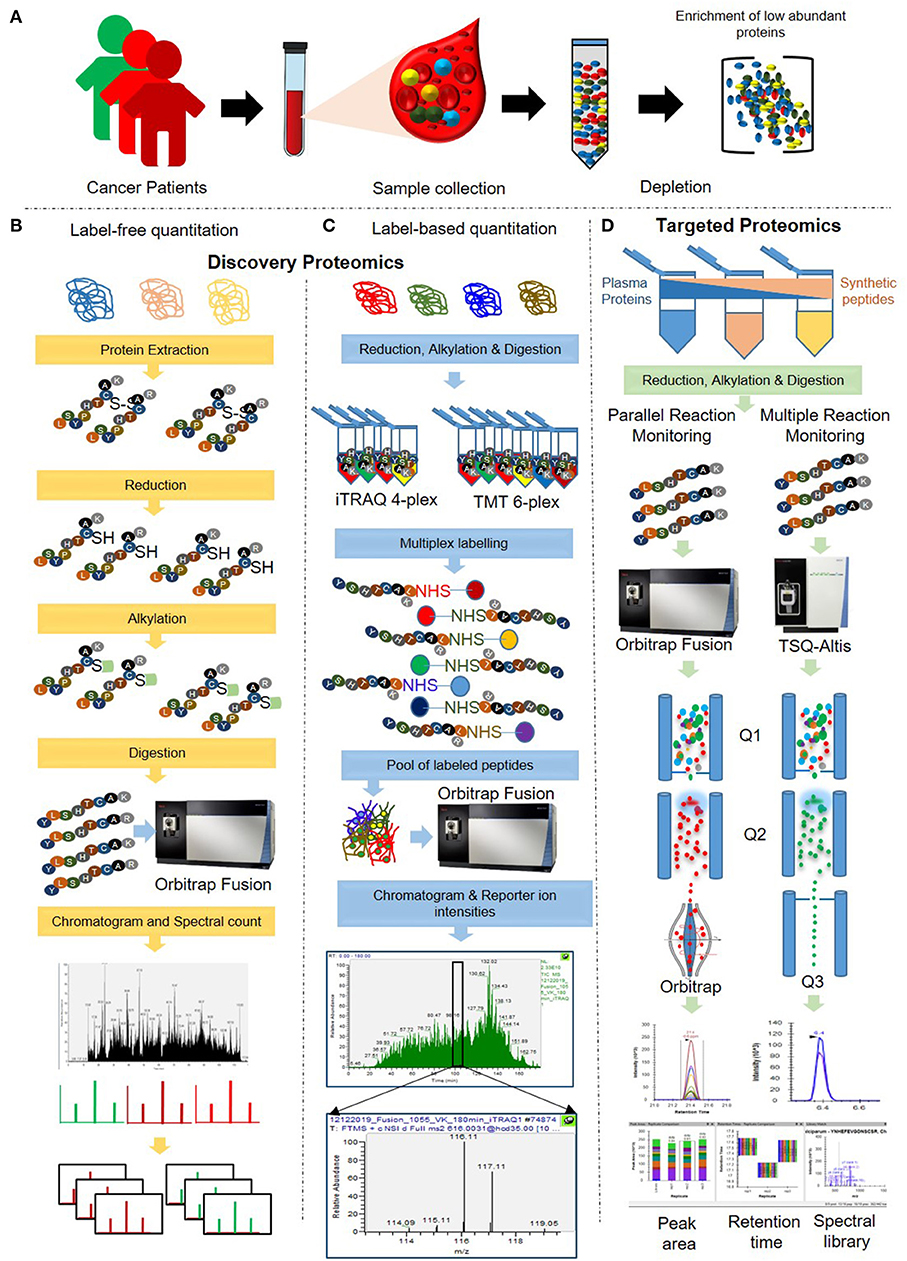
Protein Quantitation | Bruker dia-PASEF®quant outcome (Processing with Biognosys's Spectronaut™) Protein level quantitation results for a label free experiment on a 3-proteome digest. More than 8500 proteins covering 5 orders of magnitude in concentration could be identified and quantified. Advanced proteomics processing software MaxQuant data processing MaxQuant label-free quantification - Google Groups MaxQuant label-free quantification. 56 views. ... When I use MaxQuant for the LFQ analysis. I found some proteins from my experimental sample have high intensities and several peptides. Their intensities are 0 in the control sample. I am very interested in those proteins. But their LFQ intensities are 0 in both samples. Comparative evaluation of label-free quantification strategies To compare the performance of widely used or recently developed label-free quantification strategies, we analyzed the data using seven distinct label-free quantification strategies ( Fig. 1 and Supplementary Table S1). MQ-SC and StPeter were based on spectral count, while other strategies were based on ion intensity.
Maxquant label free quantification. Comparative evaluation of label-free quantification strategies The selection of a data processing method for use in mass spectrometry-based label-free proteome quantification contributes significantly to its accuracy and precision. In this study, we comprehensively evaluated 7 commonly-used label-free quantification methods (MaxQuant-Spectrum count, MaxQuant-iB … PDF Proteome-wide label-free quantification with MaxQuant - BSPR Label-free quantification Benchmark dataset •HeLa and E. coli cell lysates are mixed •Proteins were digested with trypsin. •In three replicates peptides were separated by isoelectric focusing in 24 fractions. •This was repeated with the same amount of HeLa, but E. coli lysate tripled. MaxQuant error at label free normalization step - Google Groups Then no issue with maxquant search. Good luck ... I am having a similar problem, but under my description it just says the file folder and "label free_normalization 0 Label free_normalization" and then it cuts off. I'm searching 180 files and it keeps happening. All files open fine in xcalibur, and most have searched fine in smaller searches. Plant Phosphopeptide Identification and Label-Free Quantification by ... Phosphopeptide(s) identification and label-free quantification can determine dynamic changes of phosphorylation events in plants. Both MaxQuant and Proteome Discoverer are professional software tools used to identify and quantify large-scale MS-based phosphoproteomic data.
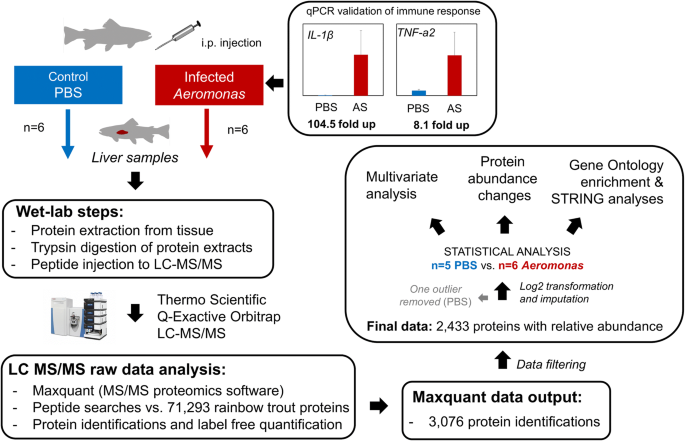
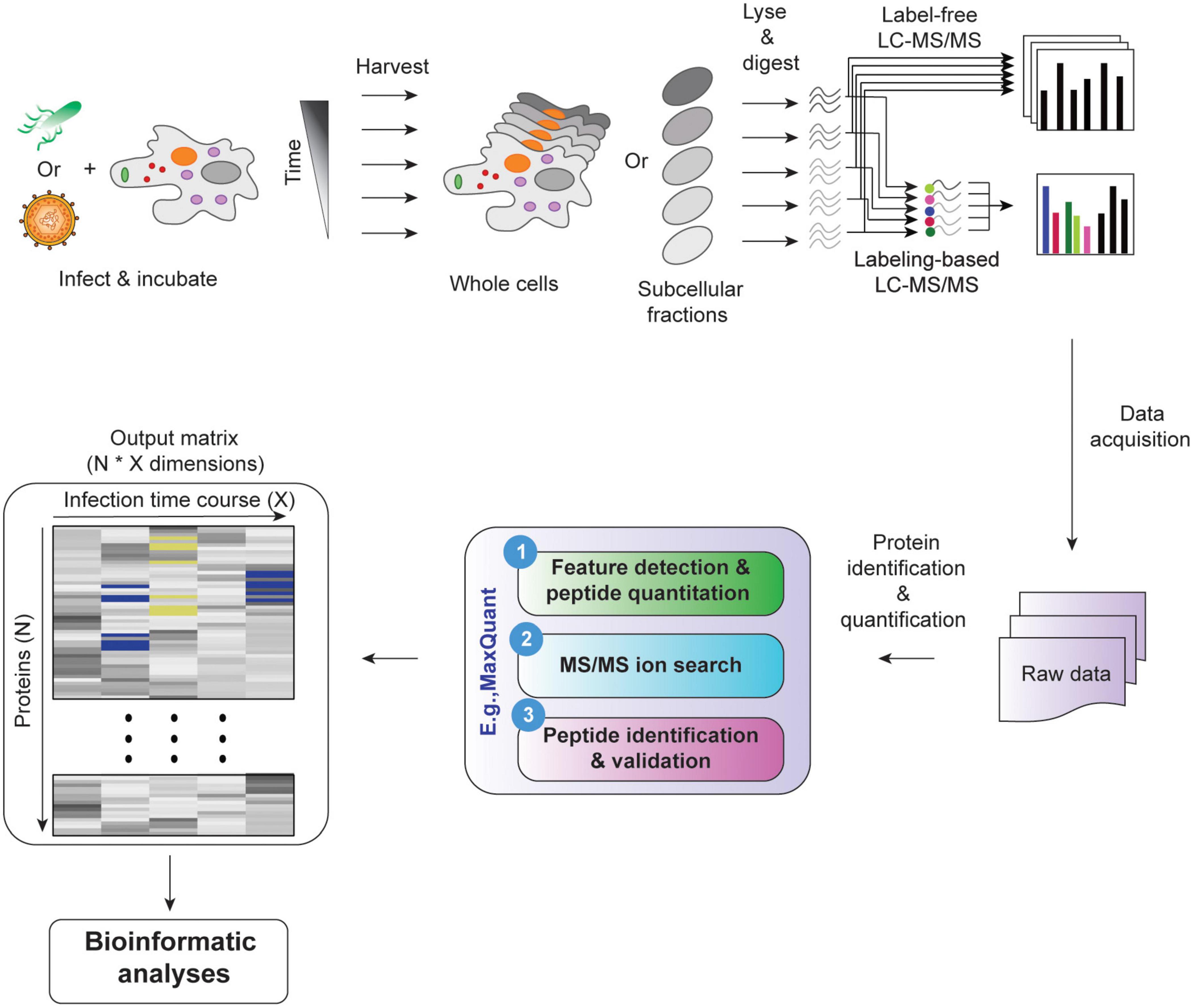
Surface proteomics and label-free quantification of Leptospira ... - PLOS Protein identification and label-free quantification by MaxQuant. Protein identification and label-free quantitative analysis were performed using MaxQuant software suite version 1.6.1.0 and its built-in Andromeda search engine. The mass spectra data (raw file format) derived from LC-MS/MS was searched against the whole L. 80 questions with answers in MAXQUANT | Science topic - ResearchGate From the Maxquant combined/text folder you need to get the info for all peptides assigned to the identified proteins from the evidence.txt file where you need to perform a lot of sorting and... MQSS 2019 | L4: Label free quantification | Christoph Wichmann To address the drawbacks and make use of the advantages of the label-free approach, MaxQuant contains a label-free quantification module called MaxLFQ. It is a series of algorithms that... What is the best experimental design in Max Quant for Label Free ... I am using MaxQuant to analyze some label-free experiment data. I did an affinity chromatography experiment and I have a technical triplicate of the elution and a technical triplicate of the...
Comparative evaluation of label-free quantification strategies To compare the performance of widely used or recently developed label-free quantification strategies, we analyzed the data using seven distinct label-free quantification strategies ( Fig. 1 and Supplementary Table S1). MQ-SC and StPeter were based on spectral count, while other strategies were based on ion intensity. MaxQuant label-free quantification - Google Groups MaxQuant label-free quantification. 56 views. ... When I use MaxQuant for the LFQ analysis. I found some proteins from my experimental sample have high intensities and several peptides. Their intensities are 0 in the control sample. I am very interested in those proteins. But their LFQ intensities are 0 in both samples. Protein Quantitation | Bruker dia-PASEF®quant outcome (Processing with Biognosys's Spectronaut™) Protein level quantitation results for a label free experiment on a 3-proteome digest. More than 8500 proteins covering 5 orders of magnitude in concentration could be identified and quantified. Advanced proteomics processing software MaxQuant data processing



![PDF] Stoichiometry of chromatin-associated protein complexes ...](https://d3i71xaburhd42.cloudfront.net/9def42dd2a83f9a86ddfebfaba77269e9d668a4d/3-Figure1-1.png)



















0 Response to "43 maxquant label free quantification"
Post a Comment